DNA-origami templates program viral coat proteins to assemble capsids with defined shapes, delivering nuclease-resistant, serum-stable cargo protection—enabling rapid, on-demand design of nanocarriers for delivery, display, and analytics without heavy protein engineering.
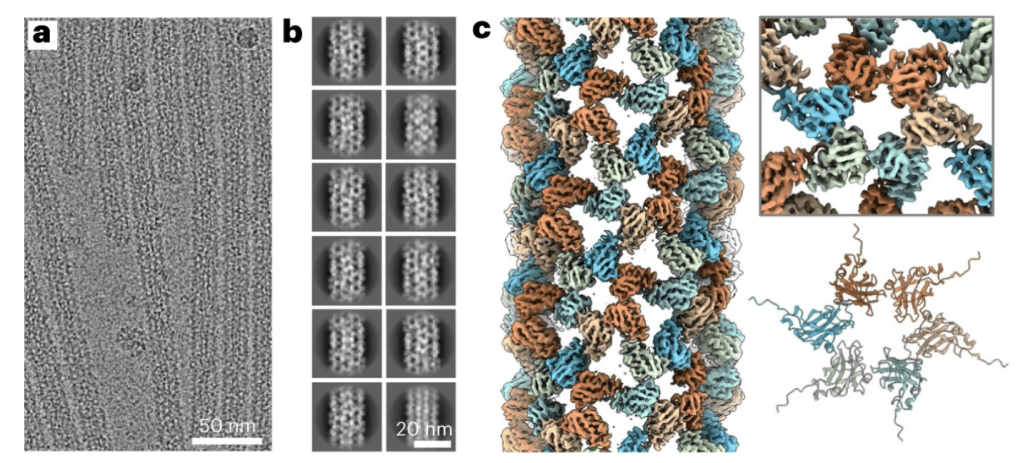
Figure from Steitz et al. (2023). License CC BY 4.0
What we studied
We evaluated a modular strategy where user-defined DNA-origami nanostructures act as assembly platforms for single-protein capsids. By templating coat proteins onto 6-helix and 24-helix DNA-origami bundles (and RNA–DNA hybrids), the team directed capsid geometry and tested whether the resulting protein coatings protect the cargo.
Key results
- Programmable morphology: Origami templates controlled capsid shape, size, and topology, yielding defined polymorphs rather than uncontrolled mixtures.
- Cargo protection: The capsid coatings shielded encapsulated origami from DNase degradation and remained stable in serum-containing media.
- Versatile materials: The approach generalized across different capsid proteins (NoV, SV40, MPyV VP1) and to RNA–DNA hybrid origami, broadening design space for therapeutics.
Why it matters
For biodesign teams building VLPs and nanocarriers, this demonstrates on-demand capsid engineering around a chosen cargo, with built-in nuclease protection. It enables rapid exploration of geometry–function relationships (e.g., cellular uptake, trafficking, antigen display) before committing to hard-to-reverse protein engineering programs.
Methods in brief
DNA/RNA–DNA origami design and folding; disassembly/reassembly of viral capsid proteins onto origami templates; negative-stain TEM and AFM; cryo-EM and helical reconstruction; biochemical stability assays in DNase and serum.
References
Seitz I., Saarinen S., Kumpula E.-P., et al. DNA-origami-directed virus capsid polymorphism. Nature Nanotechnology 18, 1205–1212 (2023). https://doi.org/10.1038/s41565-023-01443-x
